Overview
Key Features
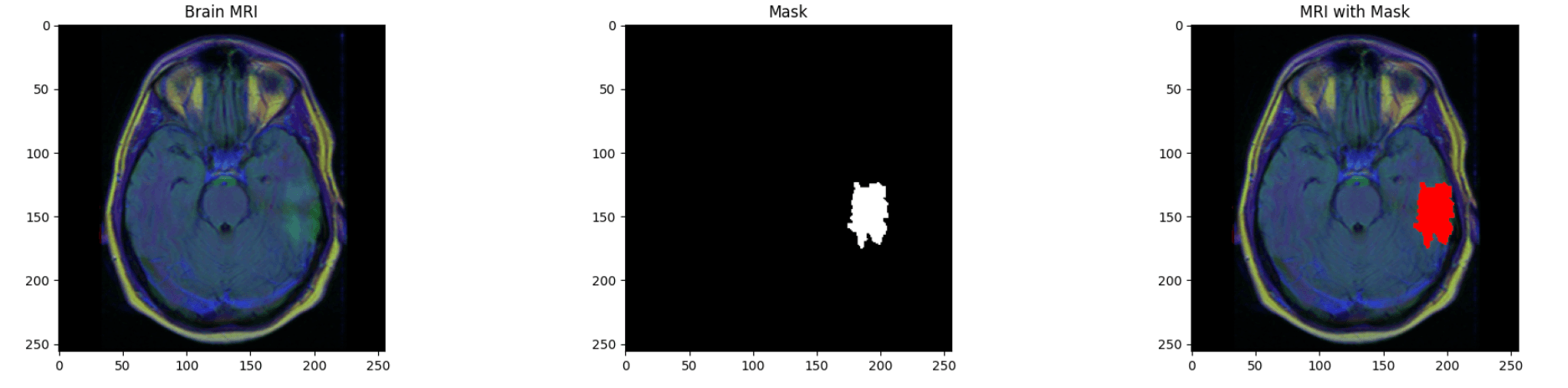
- Automated brain tumor detection in MRI scans
- Precise tumor segmentation for positive cases
- Custom data generator for efficient processing of medical imaging data
- Implementation of specialized loss functions for handling imbalanced datasets
- Integration of classification and segmentation models for comprehensive analysis
Technical Details
- Programming Language: Python
- Deep Learning Framework: TensorFlow and Keras
- Image Processing Libraries: OpenCV and scikit-image
- Data Manipulation: NumPy and Pandas
- Data Visualization: Matplotlib and Seaborn
Project Structure
brain-tumor-cnn/
│
├── data/
│ ├── data.csv
│ └── data_mask.csv
│
├── models/
│ ├── ResUNet-MRI.json
│ ├── ResUNet-model.json
│ └── resnet-50-MRI.json
│
├── notebooks/
│ └── test.ipynb
│
├── src/
│ └── utilities.py
│
├── .gitignore
├── LICENSE
├── README.md
│
└── __pycache__/Data Preparation
- The project uses a custom DataGenerator class to efficiently load and preprocess MRI images
- Data augmentation techniques are applied to increase the diversity of the training set
- Images are resized to 256x256 pixels and normalized
Model Architecture
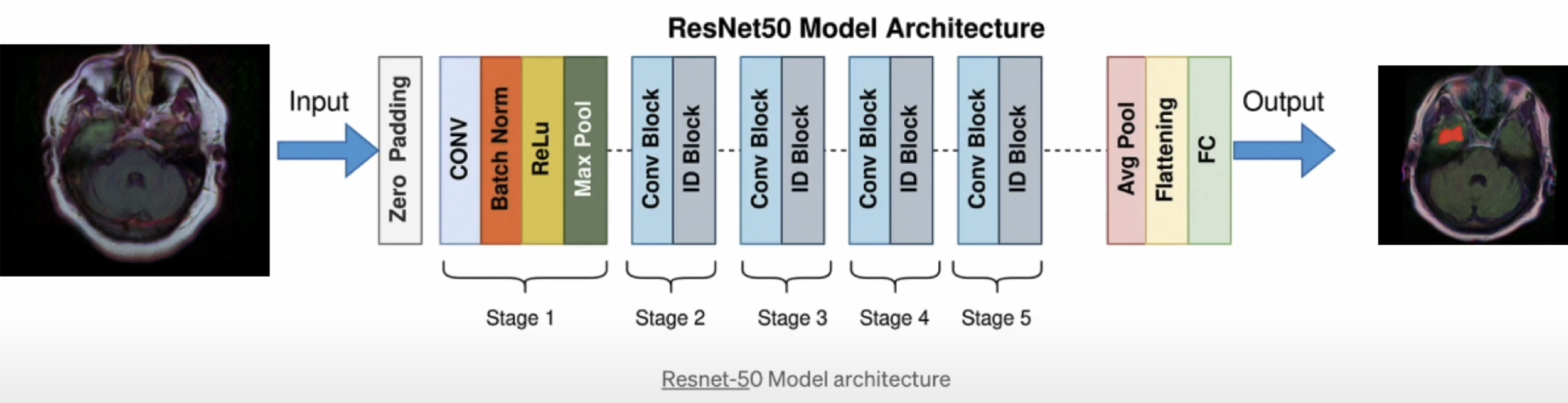
- Classification Model: ResNet-50
- Segmentation Model: ResUNet
ResNet-50 is a specific architecture of Residual Networks (ResNet), which are deep convolutional neural networks designed to address the vanishing gradient problem in very deep networks.
Key features of ResNet-50 include:
i. Depth: ResNet-50 has 49 layers, including 36 convolutional layers and 13 residual blocks.
ii. Residual Connections: ResNet-50 uses residual connections to help the network learn more complex features and avoid the vanishing gradient problem.
iii. Pre-trained on ImageNet: ResNet-50 was pre-trained on the ImageNet dataset, which contains over 14 million images and 1000 categories.
iv. Fine-tuning: ResNet-50 can be fine-tuned for specific tasks, such as tumor detection in medical images.
Pre-trained on ImageNet and fine-tuned for tumor detection.
The goal was to categorize medical images including MRI scans and X-rays into one of two cases:
i. The tumor is present
ii. The tumor is absent (normal)
ResUNet is a combination of ResNet and U-Net architectures, designed for image segmentation tasks.
Key features of ResUNet include:
i. Combines residual learning from ResNet with the U-shaped architecture of U-Net.
ii. Encoder-decoder structure with skip connections
iii. Particularly effective for medical image segmentation
iv. Maintains high-resolution features throughout the network
In this project, ResUNet is used for the segmentation task (outlining tumor regions)
Training the model
- The models were trained using a two-stage approach:
- Custom loss functions (Tversky and Focal Tversky) are implemented to handle class imbalance in segmentation tasks.
i. The classification model is trained to detect the presence of tumors.
ii. The segmentation model is trained on positive cases to outline tumor regions.
Evaluation
i. Classification performance is evaluated using accuracy, precision, recall, and F1-score.
ii. Segmentation performance is assessed using Dice coefficient and Intersection over Union (IoU).
iii. A comprehensive evaluation pipeline combines both models to provide end-to-end tumor detection and segmentation.
Plans for the future
i. Implement 3D convolutions to better utilize volumetric MRI data.
ii. Explore ensemble methods for improved classification accuracy.
iii. Investigate attention mechanisms for enhanced segmentation performance.
iv. Develop a user-friendly web interface for easy model deployment.
Results